
In the previous notebook, we introduced Pandas, which provides high-level data structures and functions designed to make working with structured or tabular data fast, easy, and expressive.
In this notebook, we will build on our knowledge of Pandas to be more productive. Pandas provides sophisticated, multi-level indexing functionality, along with the ability to perform data aggregation operations, such as grouping, merging, and joining data. It also provides capabilities for working with Time Series data that involves navigating and manipulating various date ranges and time indices. Let's dive into the details.
# Import libraries
import pandas as pd
import numpy as npWorking with Categorical Data
In many practical Data Science activities, you may come across data that contain categorical variables. These variables are typically stored as text values in columns. For such data, you may want to find the unique elements, frequency of each category present, or transform the categorical data into suitable numeric values.
Pandas provides various approaches to handle categorical data. To get started, let's create a small dataset and look at some examples. Seaborn library comes preloaded with some sample datasets. We will load the tips data from seaborn for our analysis.
import seaborn as sns
tips_data = sns.load_dataset('tips')
print(tips_data.head())total_bill tip sex smoker day time size 0 16.99 1.01 Female No Sun Dinner 2 1 10.34 1.66 Male No Sun Dinner 3 2 21.01 3.50 Male No Sun Dinner 3 3 23.68 3.31 Male No Sun Dinner 2 4 24.59 3.61 Female No Sun Dinner 4
Unique Values and Value Counts
Pandas provides methods such as unique(), nunique(), and value_counts() to extract information about the values in a column.
unique() and nunique()
unique() can be used to identify the unique elements of a column.
tips_data['day'].unique()[Sun, Sat, Thur, Fri] Categories (4, object): [Sun, Sat, Thur, Fri]
The result is an array which can be easily converted to a list by chaining the tolist() function.
tips_data['day'].unique().tolist()['Sun', 'Sat', 'Thur', 'Fri']
Similarly, unique() can be applied on the index.
indexed_tip = tips_data.set_index('day')
indexed_tip.head()| total_bill | tip | sex | smoker | time | size | |
|---|---|---|---|---|---|---|
| day | ||||||
| Sun | 16.99 | 1.01 | Female | No | Dinner | 2 |
| Sun | 10.34 | 1.66 | Male | No | Dinner | 3 |
| Sun | 21.01 | 3.50 | Male | No | Dinner | 3 |
| Sun | 23.68 | 3.31 | Male | No | Dinner | 2 |
| Sun | 24.59 | 3.61 | Female | No | Dinner | 4 |
indexed_tip.index.unique()CategoricalIndex(['Sun', 'Sat', 'Thur', 'Fri'], categories=['Sun', 'Sat', 'Thur', 'Fri'], ordered=False, name='day', dtype='category')
nunique() can be used to count the number of unique values in a column.
# Count of unique values in employee
tips_data['day'].nunique()4
# Count of unique values in skills
tips_data['time'].nunique()2
value_counts()
value_counts() are used to determine the frequency of different values present in the column.
tips_data['day'].value_counts()Sat 87 Sun 76 Thur 62 Fri 19 Name: day, dtype: int64
reset_index() can be chained to the value_counts() operation to easily get the results as a DataFrame.
days_df = tips_data['day'].value_counts().reset_index()
days_df| index | day | |
|---|---|---|
| 0 | Sat | 87 |
| 1 | Sun | 76 |
| 2 | Thur | 62 |
| 3 | Fri | 19 |
One Hot Encoding
Many machine learning algorithms do not support the presence of categorical values in data. Pandas provides various approaches to transform the categorical data into suitable numeric values to create dummy variables, and one such approach is called One Hot Encoding. The basic strategy is to convert each category value into a new column and assign a 0 or 1 (True/False) value to the column. Dummy variables can be created using get_dummies.
pd.get_dummies(tips_data, columns=['day'])| total_bill | tip | sex | smoker | time | size | day_Thur | day_Fri | day_Sat | day_Sun | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 1.01 | Female | No | Dinner | 2 | 0 | 0 | 0 | 1 |
| 1 | 10.34 | 1.66 | Male | No | Dinner | 3 | 0 | 0 | 0 | 1 |
| 2 | 21.01 | 3.50 | Male | No | Dinner | 3 | 0 | 0 | 0 | 1 |
| 3 | 23.68 | 3.31 | Male | No | Dinner | 2 | 0 | 0 | 0 | 1 |
| 4 | 24.59 | 3.61 | Female | No | Dinner | 4 | 0 | 0 | 0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 239 | 29.03 | 5.92 | Male | No | Dinner | 3 | 0 | 0 | 1 | 0 |
| 240 | 27.18 | 2.00 | Female | Yes | Dinner | 2 | 0 | 0 | 1 | 0 |
| 241 | 22.67 | 2.00 | Male | Yes | Dinner | 2 | 0 | 0 | 1 | 0 |
| 242 | 17.82 | 1.75 | Male | No | Dinner | 2 | 0 | 0 | 1 | 0 |
| 243 | 18.78 | 3.00 | Female | No | Dinner | 2 | 1 | 0 | 0 | 0 |
244 rows × 10 columns
The resulting dataset contains four new columns (one for each day) day_Thur, day_Fri, day_Sat, day_Sun. You can pass as many category columns as you would like and choose how to label the columns using prefix parameter.
pd.get_dummies(tips_data, columns=['day', 'smoker', 'sex'], prefix=['weekday', 'smokes', 'gender'])| total_bill | tip | time | size | weekday_Thur | weekday_Fri | weekday_Sat | weekday_Sun | smokes_Yes | smokes_No | gender_Male | gender_Female | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 1.01 | Dinner | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 |
| 1 | 10.34 | 1.66 | Dinner | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 |
| 2 | 21.01 | 3.50 | Dinner | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 |
| 3 | 23.68 | 3.31 | Dinner | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 |
| 4 | 24.59 | 3.61 | Dinner | 4 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 239 | 29.03 | 5.92 | Dinner | 3 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| 240 | 27.18 | 2.00 | Dinner | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 |
| 241 | 22.67 | 2.00 | Dinner | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| 242 | 17.82 | 1.75 | Dinner | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
| 243 | 18.78 | 3.00 | Dinner | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
244 rows × 12 columns
Binning Continous Variables
You may come across scenarios where you need to bin continuous data into discrete chunks to be used as a categorical variable. We can use pd.cut() function to cut our data into discrete buckets.
# Bin data into 5 equal sized buckets
pd.cut(tips_data['total_bill'], bins=5)0 (12.618, 22.166]
1 (3.022, 12.618]
2 (12.618, 22.166]
3 (22.166, 31.714]
4 (22.166, 31.714]
...
239 (22.166, 31.714]
240 (22.166, 31.714]
241 (22.166, 31.714]
242 (12.618, 22.166]
243 (12.618, 22.166]
Name: total_bill, Length: 244, dtype: category
Categories (5, interval[float64]): [(3.022, 12.618] < (12.618, 22.166] < (22.166, 31.714] < (31.714, 41.262] < (41.262, 50.81]]The function results in five equal-width bins. We can also specify bin edges to create specific non-uniform bins.
# Bin data by specifying bin edges
bill_cat = pd.cut(tips_data['total_bill'], bins=[0, 31.714, 50.81])
bill_cat0 (0.0, 31.714]
1 (0.0, 31.714]
2 (0.0, 31.714]
3 (0.0, 31.714]
4 (0.0, 31.714]
...
239 (0.0, 31.714]
240 (0.0, 31.714]
241 (0.0, 31.714]
242 (0.0, 31.714]
243 (0.0, 31.714]
Name: total_bill, Length: 244, dtype: category
Categories (2, interval[float64]): [(0.0, 31.714] < (31.714, 50.81]]# Value count for each category
bill_cat.value_counts()(0.0, 31.714] 218 (31.714, 50.81] 26 Name: total_bill, dtype: int64
The operation creates two non-uniform categories for total_bill.
Data Aggregation
Summarizing data by applying various aggregation functions such as sum(), mean(), median() etc. to each group or category within the data is a critical component of a data analysis workflow. Simple aggregations can give you a high level overview but are often not enough to get a deeper understanding of the data.
Pandas provides a flexible groupby() operation which allows for quick and efficient aggregation on subsets of data.
GroupBy
The name "group by" comes from a command in the SQL language. Hadley Wickham, author of popular packages in R programming language, described grouping operations by coining the term split-apply-combine.
- The split step breaks up and groups a DataFrame based on the value of the specified key. Splitting is performed on a particular axis of a DataFrame i.e. rows (axis=0) or columns (axis=1).
- The apply step computes some aggregation function within the individual groups.
- The combine step merges the results of these operations into an output array.
The image below shows a mockup of a simple group aggregation.
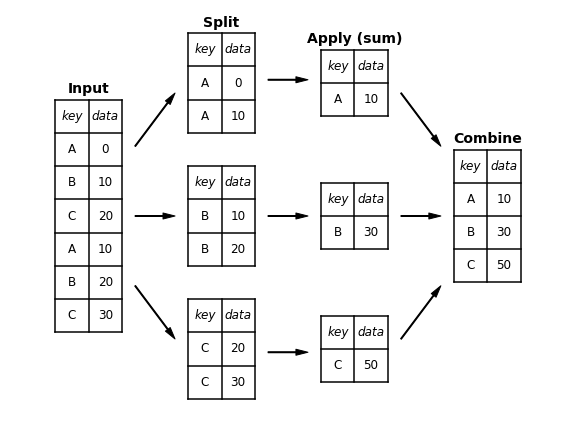
groupby() method can be used to apply the basic split-apply-combine operation on a DataFrame by specifying the desired column name.
# Apply Group By
grp1 = tips_data.groupby('sex')
grp1<pandas.core.groupby.generic.DataFrameGroupBy object at 0x7ff1186b91c0>
The method returns a DataFrameGroupBy object. No actual computation has been performed by the groupby() method yet. The idea is that this object has all the information needed to then apply some operation to each of the groups in the data. This "lazy evaluation" approach means that common aggregation functions can be implemented very efficiently using groupby(). For example, to compute the mean, we can call mean() method on the GroupBy object.
# Compute mean
grp1.mean()| total_bill | tip | size | |
|---|---|---|---|
| sex | |||
| Male | 20.744076 | 3.089618 | 2.630573 |
| Female | 18.056897 | 2.833448 | 2.459770 |
The data has been aggregated according to the group key and is now indexed by the unique values in the sex column. By default, all of the numeric columns are aggregated.
Using Multiple Keys
Multiple column names can be passed as group keys to group the data appropriately. Let's group the data by smoker and day columns.
# Aggregation using multiple keys
tips_data.groupby(['smoker', 'day']).mean()| total_bill | tip | size | ||
|---|---|---|---|---|
| smoker | day | |||
| Yes | Thur | 19.190588 | 3.030000 | 2.352941 |
| Fri | 16.813333 | 2.714000 | 2.066667 | |
| Sat | 21.276667 | 2.875476 | 2.476190 | |
| Sun | 24.120000 | 3.516842 | 2.578947 | |
| No | Thur | 17.113111 | 2.673778 | 2.488889 |
| Fri | 18.420000 | 2.812500 | 2.250000 | |
| Sat | 19.661778 | 3.102889 | 2.555556 | |
| Sun | 20.506667 | 3.167895 | 2.929825 |
The data is now indexed by the unique values in the smoker and day columns. Similarly, other aggregation operations such as sum(), median(), std() etc. can be applied to the groups within data.
# Sum operation
tips_data.groupby(['smoker', 'day']).sum()| total_bill | tip | size | ||
|---|---|---|---|---|
| smoker | day | |||
| Yes | Thur | 326.24 | 51.51 | 40 |
| Fri | 252.20 | 40.71 | 31 | |
| Sat | 893.62 | 120.77 | 104 | |
| Sun | 458.28 | 66.82 | 49 | |
| No | Thur | 770.09 | 120.32 | 112 |
| Fri | 73.68 | 11.25 | 9 | |
| Sat | 884.78 | 139.63 | 115 | |
| Sun | 1168.88 | 180.57 | 167 |
# Median operation
tips_data.groupby(['smoker', 'day']).median()| total_bill | tip | size | ||
|---|---|---|---|---|
| smoker | day | |||
| Yes | Thur | 16.470 | 2.560 | 2 |
| Fri | 13.420 | 2.500 | 2 | |
| Sat | 20.390 | 2.690 | 2 | |
| Sun | 23.100 | 3.500 | 2 | |
| No | Thur | 15.950 | 2.180 | 2 |
| Fri | 19.235 | 3.125 | 2 | |
| Sat | 17.820 | 2.750 | 2 | |
| Sun | 18.430 | 3.020 | 3 |
# Standard Deviation operation
tips_data.groupby(['smoker', 'day']).std()| total_bill | tip | size | ||
|---|---|---|---|---|
| smoker | day | |||
| Yes | Thur | 8.355149 | 1.113491 | 0.701888 |
| Fri | 9.086388 | 1.077668 | 0.593617 | |
| Sat | 10.069138 | 1.630580 | 0.862161 | |
| Sun | 10.442511 | 1.261151 | 0.901591 | |
| No | Thur | 7.721728 | 1.282964 | 1.179796 |
| Fri | 5.059282 | 0.898494 | 0.500000 | |
| Sat | 8.939181 | 1.642088 | 0.784960 | |
| Sun | 8.130189 | 1.224785 | 1.032674 |
Using aggregate()
aggregate() method allows for even greater flexibility by taking a string, a function, or a list and computing all the aggregates at once. The example below shows minimum aggregation operation being used as a string, median being called as a function, and all aggregation operations being passed as a list.
tips_data.groupby(['smoker', 'day']).aggregate(['min', np.median, max])| total_bill | tip | size | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| min | median | max | min | median | max | min | median | max | ||
| smoker | day | |||||||||
| Yes | Thur | 10.34 | 16.470 | 43.11 | 2.00 | 2.560 | 5.00 | 2 | 2 | 4 |
| Fri | 5.75 | 13.420 | 40.17 | 1.00 | 2.500 | 4.73 | 1 | 2 | 4 | |
| Sat | 3.07 | 20.390 | 50.81 | 1.00 | 2.690 | 10.00 | 1 | 2 | 5 | |
| Sun | 7.25 | 23.100 | 45.35 | 1.50 | 3.500 | 6.50 | 2 | 2 | 5 | |
| No | Thur | 7.51 | 15.950 | 41.19 | 1.25 | 2.180 | 6.70 | 1 | 2 | 6 |
| Fri | 12.46 | 19.235 | 22.75 | 1.50 | 3.125 | 3.50 | 2 | 2 | 3 | |
| Sat | 7.25 | 17.820 | 48.33 | 1.00 | 2.750 | 9.00 | 1 | 2 | 4 | |
| Sun | 8.77 | 18.430 | 48.17 | 1.01 | 3.020 | 6.00 | 2 | 3 | 6 | |
Aggregation functions can also be passed as a dictionary, mapping column names to operations that are to be applied on that column. The example below shows min operation applied to total_bill column and max operation applied to tip column.
tips_data.groupby(['smoker', 'day']).aggregate({'total_bill':'min', 'tip':'max'})| total_bill | tip | ||
|---|---|---|---|
| smoker | day | ||
| Yes | Thur | 10.34 | 5.00 |
| Fri | 5.75 | 4.73 | |
| Sat | 3.07 | 10.00 | |
| Sun | 7.25 | 6.50 | |
| No | Thur | 7.51 | 6.70 |
| Fri | 12.46 | 3.50 | |
| Sat | 7.25 | 9.00 | |
| Sun | 8.77 | 6.00 |
A more complex operation could involve passing a list of operations to be applied to a specific column.
tips_data.groupby(['smoker', 'day']).aggregate({'total_bill':['min','max','count'], 'tip':'max'})| total_bill | tip | ||||
|---|---|---|---|---|---|
| min | max | count | max | ||
| smoker | day | ||||
| Yes | Thur | 10.34 | 43.11 | 17 | 5.00 |
| Fri | 5.75 | 40.17 | 15 | 4.73 | |
| Sat | 3.07 | 50.81 | 42 | 10.00 | |
| Sun | 7.25 | 45.35 | 19 | 6.50 | |
| No | Thur | 7.51 | 41.19 | 45 | 6.70 |
| Fri | 12.46 | 22.75 | 4 | 3.50 | |
| Sat | 7.25 | 48.33 | 45 | 9.00 | |
| Sun | 8.77 | 48.17 | 57 | 6.00 | |
Selecting a Subset of Columns
For large datasets, it may be desirable to aggregate a specific column or only a subset of columns. As an example, we can group the data by smoker and compute mean for tip column as follows:
tips_data.groupby(['smoker'])['tip'].mean()smoker Yes 3.008710 No 2.991854 Name: tip, dtype: float64
Similarly, we can group the data by smoker and day columns, compute median for tip column.
tips_data.groupby(['smoker','day'])['tip'].median()smoker day
Yes Thur 2.560
Fri 2.500
Sat 2.690
Sun 3.500
No Thur 2.180
Fri 3.125
Sat 2.750
Sun 3.020
Name: tip, dtype: float64Pivot Tables
Pivot Table is a popular operation that is commonly used on tabular data in spreadsheets. The pivot table takes simple column-wise data as input, and groups the entries into a two-dimensional table that provides a multidimensional summarization of the data. Pivot Tables are essentially a multidimensional version of GroupBy. Pandas includes a pandas.pivot_table function and DataFrame also has a pivot_table method.
Seaborn library comes preloaded with some sample datasets. We will load the titanic dataset from seaborn for our analysis and look at some examples.
# Get Data
titanic_df = sns.load_dataset('titanic')
titanic_df.head()| survived | pclass | sex | age | sibsp | parch | fare | embarked | class | who | adult_male | deck | embark_town | alive | alone | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 3 | male | 22.0 | 1 | 0 | 7.2500 | S | Third | man | True | NaN | Southampton | no | False |
| 1 | 1 | 1 | female | 38.0 | 1 | 0 | 71.2833 | C | First | woman | False | C | Cherbourg | yes | False |
| 2 | 1 | 3 | female | 26.0 | 0 | 0 | 7.9250 | S | Third | woman | False | NaN | Southampton | yes | True |
| 3 | 1 | 1 | female | 35.0 | 1 | 0 | 53.1000 | S | First | woman | False | C | Southampton | yes | False |
| 4 | 0 | 3 | male | 35.0 | 0 | 0 | 8.0500 | S | Third | man | True | NaN | Southampton | no | True |
Let's say we want to look at the average number of people that survived by both sex and class. We can get the results using both GroupBy and pivot_table.
- Using
GroupBy
titanic_df.groupby(['sex', 'class'])['survived'].aggregate('mean').unstack()| class | First | Second | Third |
|---|---|---|---|
| sex | |||
| female | 0.968085 | 0.921053 | 0.500000 |
| male | 0.368852 | 0.157407 | 0.135447 |
- Using
pivot_table
titanic_df.pivot_table(values='survived', index='sex', columns='class')| class | First | Second | Third |
|---|---|---|---|
| sex | |||
| female | 0.968085 | 0.921053 | 0.500000 |
| male | 0.368852 | 0.157407 | 0.135447 |
We can see that the pivot_table approach is much more readable than the GroupBy and produces the same result. The default aggregation operation is mean.
Multi-level Pivot Table
Just as in GroupBy, the data can be grouped at multiple levels using pivot_table. Suppose we want to group survival by sex and age. Since age is a continuous variable, we can create bins for age using pd.cut function and then group the data.
# Create bins for Age
age = pd.cut(titanic_df['age'], bins=[0,18,80])
age0 (18.0, 80.0]
1 (18.0, 80.0]
2 (18.0, 80.0]
3 (18.0, 80.0]
4 (18.0, 80.0]
...
886 (18.0, 80.0]
887 (18.0, 80.0]
888 NaN
889 (18.0, 80.0]
890 (18.0, 80.0]
Name: age, Length: 891, dtype: category
Categories (2, interval[int64]): [(0, 18] < (18, 80]]# Group data
titanic_df.pivot_table(values='survived', index=['sex', age], columns='class')| class | First | Second | Third | |
|---|---|---|---|---|
| sex | age | |||
| female | (0, 18] | 0.909091 | 1.000000 | 0.511628 |
| (18, 80] | 0.972973 | 0.900000 | 0.423729 | |
| male | (0, 18] | 0.800000 | 0.600000 | 0.215686 |
| (18, 80] | 0.375000 | 0.071429 | 0.133663 |
The operation can be applied to columns in a similar fashion. Suppose we want to group survival by sex and age and look at the data by class and fare.
We can discretize the fare variable into equal-sized buckets based on sample quantiles using pd.qcut and then group the data.
# Create bins for fare
fare = pd.qcut(titanic_df['fare'], q=2)# Group data
titanic_df.pivot_table(values='survived', index=['sex', age], columns=[fare, 'class'])| fare | (-0.001, 14.454] | (14.454, 512.329] | |||||
|---|---|---|---|---|---|---|---|
| class | First | Second | Third | First | Second | Third | |
| sex | age | ||||||
| female | (0, 18] | NaN | 1.000000 | 0.714286 | 0.909091 | 1.000000 | 0.318182 |
| (18, 80] | NaN | 0.880000 | 0.444444 | 0.972973 | 0.914286 | 0.391304 | |
| male | (0, 18] | NaN | 0.000000 | 0.260870 | 0.800000 | 0.818182 | 0.178571 |
| (18, 80] | 0.0 | 0.098039 | 0.125000 | 0.391304 | 0.030303 | 0.192308 | |
Using aggfunc
aggfunc keyword can be used to specify the aggregate functions that can be applied to different columns.
titanic_df.pivot_table(index='sex', columns='class',
aggfunc={'survived':'sum', 'fare': 'mean'})| fare | survived | |||||
|---|---|---|---|---|---|---|
| class | First | Second | Third | First | Second | Third |
| sex | ||||||
| female | 106.125798 | 21.970121 | 16.118810 | 91 | 70 | 72 |
| male | 67.226127 | 19.741782 | 12.661633 | 45 | 17 | 47 |
To compute totals along each grouping, margins keyword can be used.
titanic_df.pivot_table(values='survived', index='sex', columns='class',
aggfunc='sum', margins=True)| class | First | Second | Third | All |
|---|---|---|---|---|
| sex | ||||
| female | 91 | 70 | 72 | 233 |
| male | 45 | 17 | 47 | 109 |
| All | 136 | 87 | 119 | 342 |
describe() method
Convenience methods, such as describe(), can be used to compute several common aggregates for each column. It also comes in handy when you are trying to understand the overall properties of a dataset.
tips_data.describe()| total_bill | tip | size | |
|---|---|---|---|
| count | 244.000000 | 244.000000 | 244.000000 |
| mean | 19.785943 | 2.998279 | 2.569672 |
| std | 8.902412 | 1.383638 | 0.951100 |
| min | 3.070000 | 1.000000 | 1.000000 |
| 25% | 13.347500 | 2.000000 | 2.000000 |
| 50% | 17.795000 | 2.900000 | 2.000000 |
| 75% | 24.127500 | 3.562500 | 3.000000 |
| max | 50.810000 | 10.000000 | 6.000000 |
describe() on GroupBy
describe() can be used on a groupby() object to get common aggregates for a subset of data.
tips_data.groupby(['smoker','day'])['total_bill'].describe()| count | mean | std | min | 25% | 50% | 75% | max | ||
|---|---|---|---|---|---|---|---|---|---|
| smoker | day | ||||||||
| Yes | Thur | 17.0 | 19.190588 | 8.355149 | 10.34 | 13.510 | 16.470 | 19.8100 | 43.11 |
| Fri | 15.0 | 16.813333 | 9.086388 | 5.75 | 11.690 | 13.420 | 18.6650 | 40.17 | |
| Sat | 42.0 | 21.276667 | 10.069138 | 3.07 | 13.405 | 20.390 | 26.7925 | 50.81 | |
| Sun | 19.0 | 24.120000 | 10.442511 | 7.25 | 17.165 | 23.100 | 32.3750 | 45.35 | |
| No | Thur | 45.0 | 17.113111 | 7.721728 | 7.51 | 11.690 | 15.950 | 20.2700 | 41.19 |
| Fri | 4.0 | 18.420000 | 5.059282 | 12.46 | 15.100 | 19.235 | 22.5550 | 22.75 | |
| Sat | 45.0 | 19.661778 | 8.939181 | 7.25 | 14.730 | 17.820 | 20.6500 | 48.33 | |
| Sun | 57.0 | 20.506667 | 8.130189 | 8.77 | 14.780 | 18.430 | 25.0000 | 48.17 |
Combining Data
Data science workflows often involve combining data from different sources to enhance the analysis. There are multiple ways in which data can be combined ranging from the straightforward concatenation of two different datasets, to more complicated database-style joins.
- pandas.concat - stacks together objects along an axis.
- DataFrame.append - works similar to
pandas.concatbut does not modify the original object. It creates a new object with the combined data. - pandas.merge - joins rows in DataFrame based on one or more keys. It works as the entry point for all standard database join operations between
DataFrameorSeriesobjects. - DataFrame.join -
joininstance of a DataFrame can be used for merging by index. It can be used to combine many DataFrame objects with same or similar indexes but non-overlapping columns.
concat()
concat() can be used to stack data frames together along an axis.
# Create Data
df2 = pd.DataFrame(np.arange(6).reshape(3, 2), index=['a', 'b', 'c'],
columns=['one', 'two'])
print('df2 is: \n', df2)
df3 = pd.DataFrame(5 + np.arange(4).reshape(2, 2), index=['a', 'c'],
columns=['three', 'four'])
print('\ndf3 is: \n', df3)
df4 = pd.DataFrame(5 + np.arange(4).reshape(2, 2), index=['a', 'c'],
columns=['one', 'two'])
print('\ndf4 is: \n', df4)df2 is:
one two
a 0 1
b 2 3
c 4 5
df3 is:
three four
a 5 6
c 7 8
df4 is:
one two
a 5 6
c 7 8
By default, concat() works row-wise within the DataFrame (along axis=0).
pd.concat([df2,df4])| one | two | |
|---|---|---|
| a | 0 | 1 |
| b | 2 | 3 |
| c | 4 | 5 |
| a | 5 | 6 |
| c | 7 | 8 |
ignore_index flag can be used to ignore the index when it is not necessary.
pd.concat([df2,df4], ignore_index=True)| one | two | |
|---|---|---|
| 0 | 0 | 1 |
| 1 | 2 | 3 |
| 2 | 4 | 5 |
| 3 | 5 | 6 |
| 4 | 7 | 8 |
You an specify an axis along which the concatenation should take place. If axis parameter is not specified, the concatenation works row-wise generating NaN values for unmatched columns.
pd.concat([df2,df3])| one | two | three | four | |
|---|---|---|---|---|
| a | 0.0 | 1.0 | NaN | NaN |
| b | 2.0 | 3.0 | NaN | NaN |
| c | 4.0 | 5.0 | NaN | NaN |
| a | NaN | NaN | 5.0 | 6.0 |
| c | NaN | NaN | 7.0 | 8.0 |
When axis='columns' is specified, the concatenation works along columns.
pd.concat([df2,df3], axis='columns')| one | two | three | four | |
|---|---|---|---|---|
| a | 0 | 1 | 5.0 | 6.0 |
| b | 2 | 3 | NaN | NaN |
| c | 4 | 5 | 7.0 | 8.0 |
append()
append() works similar to concat() but does not modify the original object. It creates a new object with the combined data. The method works row-wise within the DataFrame (along axis=0). This method is not very efficient, as it involves the creation of a new index and data buffer.
df2.append(df4)| one | two | |
|---|---|---|
| a | 0 | 1 |
| b | 2 | 3 |
| c | 4 | 5 |
| a | 5 | 6 |
| c | 7 | 8 |
df2.append(df3)| one | two | three | four | |
|---|---|---|---|---|
| a | 0.0 | 1.0 | NaN | NaN |
| b | 2.0 | 3.0 | NaN | NaN |
| c | 4.0 | 5.0 | NaN | NaN |
| a | NaN | NaN | 5.0 | 6.0 |
| c | NaN | NaN | 7.0 | 8.0 |
merge()
merge() joins rows in DataFrame based on one or more keys. It works as the entry point for all standard database join operations. Let's create sample data and look at some examples.
# Create data
dept_df = pd.DataFrame({'employee': ['John', 'Jake', 'Jane', 'Suzi', 'Chad'],
'group': ['Accounting', 'Engineering', 'Engineering', 'Management', 'Marketing']})
print('dept_df is: \n', dept_df)
skills_df = pd.DataFrame({'group': ['Accounting', 'Accounting',
'Engineering', 'Engineering', 'Management', 'Management', 'Operations'],
'skills': ['math', 'spreadsheets', 'coding', 'linux',
'spreadsheets', 'organization', 'SAP']})
print('\nskills_df is: \n', skills_df)dept_df is:
employee group
0 John Accounting
1 Jake Engineering
2 Jane Engineering
3 Suzi Management
4 Chad Marketing
skills_df is:
group skills
0 Accounting math
1 Accounting spreadsheets
2 Engineering coding
3 Engineering linux
4 Management spreadsheets
5 Management organization
6 Operations SAP
# Apply merge
pd.merge(dept_df,skills_df)| employee | group | skills | |
|---|---|---|---|
| 0 | John | Accounting | math |
| 1 | John | Accounting | spreadsheets |
| 2 | Jake | Engineering | coding |
| 3 | Jake | Engineering | linux |
| 4 | Jane | Engineering | coding |
| 5 | Jane | Engineering | linux |
| 6 | Suzi | Management | spreadsheets |
| 7 | Suzi | Management | organization |
Notice that a column to join the data was not specified. merge() uses the overlapping column names as keys for joining data. It is a good practice to explicitly specify the column to join the data using on keyword. 'Marketing' and 'Operations' values and associated data are missing from the result as the operation returns only common set.
pd.merge(dept_df,skills_df, on='group')| employee | group | skills | |
|---|---|---|---|
| 0 | John | Accounting | math |
| 1 | John | Accounting | spreadsheets |
| 2 | Jake | Engineering | coding |
| 3 | Jake | Engineering | linux |
| 4 | Jane | Engineering | coding |
| 5 | Jane | Engineering | linux |
| 6 | Suzi | Management | spreadsheets |
| 7 | Suzi | Management | organization |
If the column names are different in the DataFrame, then left_on and right_on keywords can be used.
# Create data
emp_df = pd.DataFrame({'name': ['John', 'Jake', 'Jane', 'Suzi', 'Chad'],
'salary': [70000, 80000, 120000, 65000, 90000]})
emp_df| name | salary | |
|---|---|---|
| 0 | John | 70000 |
| 1 | Jake | 80000 |
| 2 | Jane | 120000 |
| 3 | Suzi | 65000 |
| 4 | Chad | 90000 |
# Merge
pd.merge(dept_df, emp_df, left_on='employee', right_on='name')| employee | group | name | salary | |
|---|---|---|---|---|
| 0 | John | Accounting | John | 70000 |
| 1 | Jake | Engineering | Jake | 80000 |
| 2 | Jane | Engineering | Jane | 120000 |
| 3 | Suzi | Management | Suzi | 65000 |
| 4 | Chad | Marketing | Chad | 90000 |
The redundant column can be dropped as needed using the drop() method.
pd.merge(dept_df, emp_df, left_on='employee', right_on='name').drop('name', axis='columns')| employee | group | salary | |
|---|---|---|---|
| 0 | John | Accounting | 70000 |
| 1 | Jake | Engineering | 80000 |
| 2 | Jane | Engineering | 120000 |
| 3 | Suzi | Management | 65000 |
| 4 | Chad | Marketing | 90000 |
Inner Join
By default merge() performs an inner join. The result is an intersection, or the common set found in both DataFrame. The merge operations we just saw were all inner joins. Different join types such as left, right, outer can be specified using the how= parameter.
pd.merge(dept_df,skills_df, on='group', how='inner')| employee | group | skills | |
|---|---|---|---|
| 0 | John | Accounting | math |
| 1 | John | Accounting | spreadsheets |
| 2 | Jake | Engineering | coding |
| 3 | Jake | Engineering | linux |
| 4 | Jane | Engineering | coding |
| 5 | Jane | Engineering | linux |
| 6 | Suzi | Management | spreadsheets |
| 7 | Suzi | Management | organization |
'Marketing' and 'Operations' values and associated data are missing from the result, as the operation returns only common set.
Left Join
This join returns all records from the left DataFrame and the matched records from the right DataFrame.
pd.merge(dept_df,skills_df, on='group', how='left')| employee | group | skills | |
|---|---|---|---|
| 0 | John | Accounting | math |
| 1 | John | Accounting | spreadsheets |
| 2 | Jake | Engineering | coding |
| 3 | Jake | Engineering | linux |
| 4 | Jane | Engineering | coding |
| 5 | Jane | Engineering | linux |
| 6 | Suzi | Management | spreadsheets |
| 7 | Suzi | Management | organization |
| 8 | Chad | Marketing | NaN |
The result is a Cartesian product of the rows. Since there were two 'Engineering' rows in the left DataFrame and two in the right, there are four 'Engineering' rows in the result. Similarly, there was one 'Accounting' row in left DataFrame and two in the right, resulting in two 'Accounting' rows. The row for 'Operations' is missing as this join only keeps matched rows from right DataFrame.
Right Join
This join returns all records from the right DataFrame and the matched records from the left DataFrame.
pd.merge(dept_df,skills_df, on='group', how='right')| employee | group | skills | |
|---|---|---|---|
| 0 | John | Accounting | math |
| 1 | John | Accounting | spreadsheets |
| 2 | Jake | Engineering | coding |
| 3 | Jane | Engineering | coding |
| 4 | Jake | Engineering | linux |
| 5 | Jane | Engineering | linux |
| 6 | Suzi | Management | spreadsheets |
| 7 | Suzi | Management | organization |
| 8 | NaN | Operations | SAP |
The result is a Cartesian product of the rows. Since there were two 'Engineering' rows in the right DataFrame and two in the left, there are four 'Engineering' rows in the result. Similarly, there was one 'Accounting' row in left DataFrame and two in the right, resulting in two 'Accounting' rows. The row for 'Marketing' is missing as this join only keeps matched rows from right DataFrame.
Outer Join
The outer join takes the union of the keys, combining the effect of applying both left and right joins.
out_df = pd.merge(dept_df,skills_df, on='group', how='outer')
out_df| employee | group | skills | |
|---|---|---|---|
| 0 | John | Accounting | math |
| 1 | John | Accounting | spreadsheets |
| 2 | Jake | Engineering | coding |
| 3 | Jake | Engineering | linux |
| 4 | Jane | Engineering | coding |
| 5 | Jane | Engineering | linux |
| 6 | Suzi | Management | spreadsheets |
| 7 | Suzi | Management | organization |
| 8 | Chad | Marketing | NaN |
| 9 | NaN | Operations | SAP |
The result is a Cartesian product of the rows using all key combinations filling in all missing values with NAs.
Using index to merge
Index can also be used as the key for merging by specifying the left_index and/or right_index flags.
# Create data
dept_dfa = dept_df.set_index('group')
print('dept_dfa is \n', dept_dfa)
skills_dfa = skills_df.set_index('group')
print('\nskills_dfa is \n', skills_dfa)dept_dfa is
employee
group
Accounting John
Engineering Jake
Engineering Jane
Management Suzi
Marketing Chad
skills_dfa is
skills
group
Accounting math
Accounting spreadsheets
Engineering coding
Engineering linux
Management spreadsheets
Management organization
Operations SAP
# Merge
pd.merge(dept_dfa, skills_dfa, left_index=True, right_index=True)| employee | skills | |
|---|---|---|
| group | ||
| Accounting | John | math |
| Accounting | John | spreadsheets |
| Engineering | Jake | coding |
| Engineering | Jake | linux |
| Engineering | Jane | coding |
| Engineering | Jane | linux |
| Management | Suzi | spreadsheets |
| Management | Suzi | organization |
join()
join instance of a DataFrame can also be used for merging by index. The how keword can be specified for the type of join.
dept_dfa.join(skills_dfa, how='inner')| employee | skills | |
|---|---|---|
| group | ||
| Accounting | John | math |
| Accounting | John | spreadsheets |
| Engineering | Jake | coding |
| Engineering | Jake | linux |
| Engineering | Jane | coding |
| Engineering | Jane | linux |
| Management | Suzi | spreadsheets |
| Management | Suzi | organization |
'Marketing' and 'Operations' values and associated data are missing from the result, as the operation returns only common set.
dept_dfa.join(skills_dfa, how='outer')| employee | skills | |
|---|---|---|
| group | ||
| Accounting | John | math |
| Accounting | John | spreadsheets |
| Engineering | Jake | coding |
| Engineering | Jake | linux |
| Engineering | Jane | coding |
| Engineering | Jane | linux |
| Management | Suzi | spreadsheets |
| Management | Suzi | organization |
| Marketing | Chad | NaN |
| Operations | NaN | SAP |
The result shows all data, filling in the missing values with NAs.
Hierarchical Indexing
Hierarchical indexing (also known as multi-indexing) allows you to have multiple (two or more) index levels within a single index on an axis. It provides a way for representing higher dimensional data in a lower dimensional form. Let's start with a simple example, creating a series with multi-index.
Multi-indexed Series
# Create Data
index = [('CA', 2005), ('CA', 2015),
('NY', 2005), ('NY', 2015),
('TX', 2005), ('TX', 2015)]
population = [33871648, 37253956,
18976457, 19378102,
20851820, 25145561]
pop_series = pd.Series(population, index=index)
pop_series(CA, 2005) 33871648 (CA, 2015) 37253956 (NY, 2005) 18976457 (NY, 2015) 19378102 (TX, 2005) 20851820 (TX, 2015) 25145561 dtype: int64
With this indexing, you can easily index or slice the series using the index. However, what if you wanted to select all the values for 2015? The tuple-based index is essentially a multi-index and Pandas MultiIndex type allows us to create multi-level indexes. This provides us with the flexibility to perform operations on indexes easily and efficiently.
We will create a multi-indexed index for pop_series using the MultiIndex type.
new_index = pd.MultiIndex.from_tuples(index)
new_index.levelsFrozenList([['CA', 'NY', 'TX'], [2005, 2015]])
Notice that the new_index object contains multiple levels of indexing, the state names and the years. We can now reindex the pop_series to see hierarchical representation of the data.
pop_series = pop_series.reindex(new_index)
pop_seriesCA 2005 33871648
2015 37253956
NY 2005 18976457
2015 19378102
TX 2005 20851820
2015 25145561
dtype: int64Subset Selection
Subset selection of multi-indexed data is similar to what we have seen in the previous part of this guide series. Let's take a quick look.
- Select data for California
pop_series['CA']2005 33871648 2015 37253956 dtype: int64
- Select data from New York to Texas
pop_series['NY':'TX']NY 2005 18976457
2015 19378102
TX 2005 20851820
2015 25145561
dtype: int64- Select data for California and Texas
pop_series[['CA','TX']]CA 2005 33871648
2015 37253956
TX 2005 20851820
2015 25145561
dtype: int64We can now easily access the data for second index and answer our question about selecting all the values for 2015.
pop_series[:,2015]CA 37253956 NY 19378102 TX 25145561 dtype: int64
unstack() and stack()
unstack() method will convert a multi-indexed Series into a DataFrame, and naturally stack() would do the opposite.
# Unstack
pop_df = pop_series.unstack()
pop_df| 2005 | 2015 | |
|---|---|---|
| CA | 33871648 | 37253956 |
| NY | 18976457 | 19378102 |
| TX | 20851820 | 25145561 |
The result is a DataFrame where second level index (years) is converted to columns, and first level index (states) remains as the index of the DataFrame.
# Stack
pop_df.stack()CA 2005 33871648
2015 37253956
NY 2005 18976457
2015 19378102
TX 2005 20851820
2015 25145561
dtype: int64level can be specified to unstack() by a specific index level. Specifying level=0 will unstack based on the outermost index level i.e. by 'state'.
pop_series.unstack(level=0)| CA | NY | TX | |
|---|---|---|---|
| 2005 | 33871648 | 18976457 | 20851820 |
| 2015 | 37253956 | 19378102 | 25145561 |
Specifying level=1 unstacks by the inner index, in this case 'year'.
pop_series.unstack(level=1)| 2005 | 2015 | |
|---|---|---|
| CA | 33871648 | 37253956 |
| NY | 18976457 | 19378102 |
| TX | 20851820 | 25145561 |
Multi-indexed DataFrame
In a DataFrame, both rows and columns can have multiple levels of indices. Let's create some sample data and take a look.
# hierarchical indices and columns
index = pd.MultiIndex.from_product([[2019, 2018], [2, 1]],
names=['year', 'quarter'])
columns = pd.MultiIndex.from_product([['John', 'Jane', 'Ben'], ['Product A', 'Product B']],
names=['sales person', 'product'])
# mock some data
data = np.round(np.random.randn(4, 6), 1)
data[:, ::2] *= 10
data += 55
# create the DataFrame
sales_data = pd.DataFrame(data, index=index, columns=columns)
sales_data| sales person | John | Jane | Ben | ||||
|---|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B | |
| year | quarter | ||||||
| 2019 | 2 | 49.0 | 56.3 | 54.0 | 55.4 | 48.0 | 54.5 |
| 1 | 51.0 | 55.7 | 58.0 | 53.5 | 67.0 | 52.9 | |
| 2018 | 2 | 43.0 | 54.9 | 52.0 | 55.9 | 46.0 | 55.0 |
| 1 | 39.0 | 55.6 | 62.0 | 56.3 | 39.0 | 55.2 | |
sales_data is essentially four dimensional data with 'sales person', 'product', 'year' and 'quarter' as its dimensions.
Subset Selection
Since columns in a DataFrame are individual Series, the syntax used for multi-indexed Series applies to the columns.
- Select data for 'Product A' sold by John
sales_data['John','Product A']year quarter
2019 2 49.0
1 51.0
2018 2 43.0
1 39.0
Name: (John, Product A), dtype: float64- Select data for John and Jane
sales_data[['John','Jane']]| sales person | John | Jane | |||
|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | |
| year | quarter | ||||
| 2019 | 2 | 49.0 | 56.3 | 54.0 | 55.4 |
| 1 | 51.0 | 55.7 | 58.0 | 53.5 | |
| 2018 | 2 | 43.0 | 54.9 | 52.0 | 55.9 |
| 1 | 39.0 | 55.6 | 62.0 | 56.3 | |
.loc and .iloc index operators can also be used.
- Sales data for first two rows and first four columns
sales_data.iloc[:2, :4]| sales person | John | Jane | |||
|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | |
| year | quarter | ||||
| 2019 | 2 | 49.0 | 56.3 | 54.0 | 55.4 |
| 1 | 51.0 | 55.7 | 58.0 | 53.5 | |
- Jane's sales in 2019
sales_data.loc[2019,'Jane']| product | Product A | Product B |
|---|---|---|
| quarter | ||
| 2 | 54.0 | 55.4 |
| 1 | 58.0 | 53.5 |
- Jane's sales in 2019 for Product B
sales_data.loc[2019,('Jane','Product B')]quarter 2 55.4 1 53.5 Name: (Jane, Product B), dtype: float64
- Jane's sales in second quarter of 2019 for Product B
sales_data.loc[(2019,2), ('Jane','Product B')]55.4
Sorting
By Index and Level
sort_index() can be used to sort the index or levels within your data. By default, the indexing operartion is performed on the outermost index (level=0) and in ascending order.
sales_data.sort_index()| sales person | John | Jane | Ben | ||||
|---|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B | |
| year | quarter | ||||||
| 2018 | 1 | 39.0 | 55.6 | 62.0 | 56.3 | 39.0 | 55.2 |
| 2 | 43.0 | 54.9 | 52.0 | 55.9 | 46.0 | 55.0 | |
| 2019 | 1 | 51.0 | 55.7 | 58.0 | 53.5 | 67.0 | 52.9 |
| 2 | 49.0 | 56.3 | 54.0 | 55.4 | 48.0 | 54.5 | |
Specifying level=1 sorts by the inner index.
sales_data.sort_index(level=1)| sales person | John | Jane | Ben | ||||
|---|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B | |
| year | quarter | ||||||
| 2018 | 1 | 39.0 | 55.6 | 62.0 | 56.3 | 39.0 | 55.2 |
| 2019 | 1 | 51.0 | 55.7 | 58.0 | 53.5 | 67.0 | 52.9 |
| 2018 | 2 | 43.0 | 54.9 | 52.0 | 55.9 | 46.0 | 55.0 |
| 2019 | 2 | 49.0 | 56.3 | 54.0 | 55.4 | 48.0 | 54.5 |
Sorting can be applied on columns by specifying axis='columns'.
sales_data.sort_index(axis='columns')| sales person | Ben | Jane | John | ||||
|---|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B | |
| year | quarter | ||||||
| 2019 | 2 | 48.0 | 54.5 | 54.0 | 55.4 | 49.0 | 56.3 |
| 1 | 67.0 | 52.9 | 58.0 | 53.5 | 51.0 | 55.7 | |
| 2018 | 2 | 46.0 | 55.0 | 52.0 | 55.9 | 43.0 | 54.9 |
| 1 | 39.0 | 55.2 | 62.0 | 56.3 | 39.0 | 55.6 | |
sales_data.sort_index(axis='columns', level=1)| sales person | Ben | Jane | John | Ben | Jane | John | |
|---|---|---|---|---|---|---|---|
| product | Product A | Product A | Product A | Product B | Product B | Product B | |
| year | quarter | ||||||
| 2019 | 2 | 48.0 | 54.0 | 49.0 | 54.5 | 55.4 | 56.3 |
| 1 | 67.0 | 58.0 | 51.0 | 52.9 | 53.5 | 55.7 | |
| 2018 | 2 | 46.0 | 52.0 | 43.0 | 55.0 | 55.9 | 54.9 |
| 1 | 39.0 | 62.0 | 39.0 | 55.2 | 56.3 | 55.6 |
By Value
sort_values() can be used to sort the values in a DataFrame by one or more columns.
- Sort by values of quarter and then year
sales_data.sort_values(by=['quarter','year'])| sales person | John | Jane | Ben | ||||
|---|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B | |
| year | quarter | ||||||
| 2018 | 1 | 39.0 | 55.6 | 62.0 | 56.3 | 39.0 | 55.2 |
| 2019 | 1 | 51.0 | 55.7 | 58.0 | 53.5 | 67.0 | 52.9 |
| 2018 | 2 | 43.0 | 54.9 | 52.0 | 55.9 | 46.0 | 55.0 |
| 2019 | 2 | 49.0 | 56.3 | 54.0 | 55.4 | 48.0 | 54.5 |
For multi-indexed data, column label must be unique. So, the values passed to by= parameter must be a tuple with elements corresponding to each level.
- Sort by values of Product A for Ben
sales_data.sort_values(by=('Ben','Product A'))| sales person | John | Jane | Ben | ||||
|---|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B | |
| year | quarter | ||||||
| 2018 | 1 | 39.0 | 55.6 | 62.0 | 56.3 | 39.0 | 55.2 |
| 2 | 43.0 | 54.9 | 52.0 | 55.9 | 46.0 | 55.0 | |
| 2019 | 2 | 49.0 | 56.3 | 54.0 | 55.4 | 48.0 | 54.5 |
| 1 | 51.0 | 55.7 | 58.0 | 53.5 | 67.0 | 52.9 | |
Multiple columns, or a combination or column and index, can be specified by passing them as a list of tuples.
sales_data.sort_values(by=[('Jane','Product A'), ('quarter')])| sales person | John | Jane | Ben | ||||
|---|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B | |
| year | quarter | ||||||
| 2018 | 2 | 43.0 | 54.9 | 52.0 | 55.9 | 46.0 | 55.0 |
| 2019 | 2 | 49.0 | 56.3 | 54.0 | 55.4 | 48.0 | 54.5 |
| 1 | 51.0 | 55.7 | 58.0 | 53.5 | 67.0 | 52.9 | |
| 2018 | 1 | 39.0 | 55.6 | 62.0 | 56.3 | 39.0 | 55.2 |
Data Aggregations
We have seen data aggregations in a previous section in this notebook. Various aggregation functions such as sum(), mean(), median() can be applied to multi-indexed data.
# Get data
sales_data| sales person | John | Jane | Ben | ||||
|---|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B | |
| year | quarter | ||||||
| 2019 | 2 | 49.0 | 56.3 | 54.0 | 55.4 | 48.0 | 54.5 |
| 1 | 51.0 | 55.7 | 58.0 | 53.5 | 67.0 | 52.9 | |
| 2018 | 2 | 43.0 | 54.9 | 52.0 | 55.9 | 46.0 | 55.0 |
| 1 | 39.0 | 55.6 | 62.0 | 56.3 | 39.0 | 55.2 | |
- Total sales by sales person and product
sales_data.sum()sales person product
John Product A 182.0
Product B 222.5
Jane Product A 226.0
Product B 221.1
Ben Product A 200.0
Product B 217.6
dtype: float64- Total sales by year and quarter
sales_data.sum(axis='columns')year quarter
2019 2 317.2
1 338.1
2018 2 306.8
1 307.1
dtype: float64level parameter controls the subset of data to which aggregation is applied. Let's look at some examples.
- Total sales for each quarter by product and sales person
sales_data.sum(level='quarter')| sales person | John | Jane | Ben | |||
|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B |
| quarter | ||||||
| 2 | 92.0 | 111.2 | 106.0 | 111.3 | 94.0 | 109.5 |
| 1 | 90.0 | 111.3 | 120.0 | 109.8 | 106.0 | 108.1 |
- Average sales for each year by product and sales person
yearly_avg = sales_data.mean(level='year')
yearly_avg| sales person | John | Jane | Ben | |||
|---|---|---|---|---|---|---|
| product | Product A | Product B | Product A | Product B | Product A | Product B |
| year | ||||||
| 2019 | 50.0 | 56.00 | 56.0 | 54.45 | 57.5 | 53.7 |
| 2018 | 41.0 | 55.25 | 57.0 | 56.10 | 42.5 | 55.1 |
- Average sales per year by Product
yearly_avg.mean(axis=1, level='product')| product | Product A | Product B |
|---|---|---|
| year | ||
| 2019 | 54.500000 | 54.716667 |
| 2018 | 46.833333 | 55.483333 |
Conclusion
In this part of the guide series we learned about how to be more productive with Pandas. We started with Data Aggregation using groupby and pivot_table. Next, we discussed how data can be combined using concat(), append(), merge(), and join() methods. You have seen how data can be indexed at multiple levels in the Hierarchical indexing section. Here, we discussed multi-indexed Series and DataFrame, including selection, sorting, and aggregation methods. We also looked at how to look at unique values and value counts for categorical data.
In the next part of this guide series, we will explore the capabilities for working with Time Series data.
References
[1] Wes McKinney. 2017. Python for Data Analysis: Data Wrangling with Pandas, NumPy, and IPython (2nd. ed.). O'Reilly Media, Inc.
[2] Jake VanderPlas. 2016. Python Data Science Handbook: Essential Tools for Working with Data (1st. ed.). O'Reilly Media, Inc.